Quantum Convolutional Neural Network Classifier#
Code at: https://github.com/qiboteam/qibo/tree/master/examples/qcnn_classifier. Please note that scikit-learn is needed to visualize the results.
Problem overview#
This tutorial implements a simple Quantum Convolutional Neural Network (QCNN), which is a translationally invariant algorithm analogous to the classical convolutional neural network. This example demonstrates the use of the QCNN as a quantum classifier, which attempts to classify ground states of a translationally invariant quantum system, the transverse field Ising model, based on whether they are in the ordered or disordered phase. The (randomized) statevector data provided are those of a 4-qubit system. Accompanying each state is a label: +1 (disordered phase) or -1 (ordered phase).
Through the sequential reduction of entanglement, this network is able to perform classification from the final measurement of a single qubit.
Workflow of QCNN model:
Schematic of QCNN model:
Convolutional layer for 4 qubits as an example:
Pooling layer for 4 qubits as an example:
where in the above, \(R(\theta_{i,j,k}) = RZ(\theta_k) RY(\theta_j) RX(\theta_i)\):
\(U_{q_a, q_b}(\theta_{i,j,k}) = RXX(\theta_k) RYY(\theta_j) RZZ(\theta_i)\) is a two-qubit gate acting on qubits \(q_a\) and \(q_b\):
and \(R^{\dagger}(\theta_{i,j,k}) = RX(-\theta_i) RY(-\theta_j) RZ(-\theta_k)\):
How to use the QCNN class#
For more details on the QuantumCNN class, please refer to the documentation. Here we recall some of the necessary arguments when instantiating a QuantumCNN object: - nqubits (int): number of quantum bits. It should be larger than 2 for the model to make sense. - nlayers (int): number of layers of the QCNN variational ansatz. - nclasses (int): number of classes of the training set (default=2). - params: list to initialise the variational parameters (default=None).
After creating the object, one can proceed to train the model. For this, the QuantumCNN.minimize method can be used with the following arguments (refer to the documentation for more details)” - init_theta: list or numpy.array with the angles to be used in the circuit - data: the training data - labels: numpy.array containing the labels for the training data - nshots (int):number of runs of the circuit during the sampling process (default=10000) - method (string): str
‘classical optimizer for the minimization’. All methods from scipy.optimize.minmize are suported (default=‘Powell’).
QCNN Demo#
Include necessary packages:
[4]:
import numpy as np
import random
from sklearn import metrics
import qibo
from qibo.models.qcnn import QuantumCNN
qibo.set_backend("numpy")
[Qibo 0.1.13|INFO|2023-05-15 17:25:59]: Using numpy backend on /CPU:0
Load the provided data (ground states of 4-qubit TFIM in data folder) and labels:
[5]:
data = np.load('nqubits_4_data_shuffled_no0.npy')
labels = np.load('nqubits_4_labels_shuffled_no0.npy')
labels = np.transpose(np.array([labels])) # restructure to required array format
Structure of data and labels are like:
[6]:
data[-2:]
[6]:
array([[0.52745364+0.j, 0.19856967+0.j, 0.19856967+0.j, 0.16507377+0.j,
0.19856967+0.j, 0.09784837+0.j, 0.16507377+0.j, 0.19856967+0.j,
0.19856967+0.j, 0.16507377+0.j, 0.09784837+0.j, 0.19856967+0.j,
0.16507377+0.j, 0.19856967+0.j, 0.19856967+0.j, 0.52745364+0.j],
[0.67109214+0.j, 0.10384038+0.j, 0.10384038+0.j, 0.05351362+0.j,
0.10384038+0.j, 0.02786792+0.j, 0.05351362+0.j, 0.10384038+0.j,
0.10384038+0.j, 0.05351362+0.j, 0.02786792+0.j, 0.10384038+0.j,
0.05351362+0.j, 0.10384038+0.j, 0.10384038+0.j, 0.67109214+0.j]])
[7]:
labels[-2:]
[7]:
array([[ 1.],
[-1.]])
Split the data into training/test set in the ratio 60:40
[8]:
split_ind = int(len(data) * 0.6)
train_data = data[:split_ind]
test_data = data[split_ind:]
train_labels = labels[:split_ind]
test_labels = labels[split_ind:]
Initialize the QuantumCNN class:
[9]:
test = QuantumCNN(nqubits=4, nlayers=1, nclasses=2)
testcircuit = test._circuit
testcircuit.draw()
q0: ─RX─RY─RZ─RZZ─RYY─RXX─RX─RY─RZ──────────────────────────────────────── ...
q1: ─RX─RY─RZ─RZZ─RYY─RXX─RX─RY─RZ─────────────RX─RY─RZ──────────RZZ─RYY─R ...
q2: ──────────────────────RX─RY─RZ─RZZ─RYY─RXX─RX─RY─RZ─RX─RY─RZ─RZZ─RYY─R ...
q3: ──────────────────────RX─RY─RZ─RZZ─RYY─RXX─RX─RY─RZ─────────────────── ...
q0: ... ───RX─RY─RZ─RZZ─RYY─RXX─RX─RY─RZ─RX─RY─RZ─o───────────────────────
q1: ... XX─RX─RY─RZ─|───|───|─────────────────────|─RX─RY─RZ─o────────────
q2: ... XX─RX─RY─RZ─|───|───|───RX─RY─RZ──────────X─RZ─RY─RX─|────────────
q3: ... ───RX─RY─RZ─RZZ─RYY─RXX─RX─RY─RZ────────────RX─RY─RZ─X─RZ─RY─RX─M─
draw() is used to visualize the pre-constructed circuit based on input parameters for class initialization.
Initialize model parameters:
[ ]:
testbias = np.zeros(test.measured_qubits)
testangles = [random.uniform(0,2*np.pi) for _ in range(21*2)]
init_theta = np.concatenate((testbias, testangles))
Train model with optimize parameters (automatically updates model with optimized paramters at the end of training):
[ ]:
result = test.minimize(init_theta, data=train_data, labels=labels, nshots=10000, method='Powell')
Alternatively, update model with optimized parameters from previous training:
[11]:
saved_result_60 = (0.2026119742575817, np.array([ -0.06559061, 3.62881221, 2.39850148, 3.02493711,
0.91498683, 3.25517842, 0.0759049 , 3.46049453,
3.04395784, 1.55681424, 2.3665245 , 0.40291846,
5.67310744, 2.27615444, 5.23403537, 0.46053411,
0.69228362, 2.2308165 , 0.53323661, 4.52157388,
5.31194656, 18.23511858, -1.90754635, 14.30577217,
10.75135972, 19.16001316, 12.27582746, 7.47476354,
23.38129141, 60.29771502, 10.02946377, 17.83945879,
15.22732248, 12.34666584, 1.52634649, 1.90621517,
12.71554053, -13.56379057, 34.04591253, -11.56450878,
10.95038782, 3.30640208, 9.67270071]))
test.set_circuit_params(angles=saved_result_60[1], has_bias=True)
Generate predictions from optimized model:
[12]:
predictions = []
for n in range(len(test_data)):
predictions.append(test.predict(test_data[n], nshots=10000)[0])
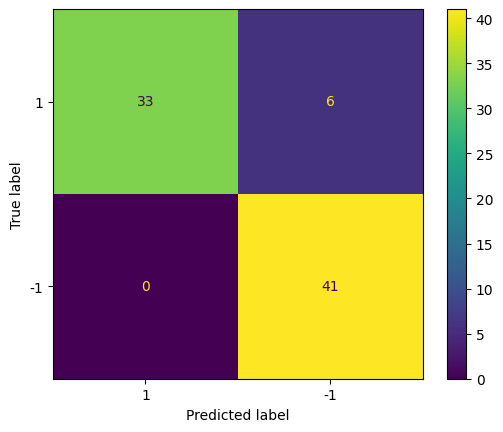
Visualize results via confusion matrix and accuracy:
[14]:
actual = [np.sign(test_labels) for test_labels in test_labels]
predicted = [np.sign(prediction) for prediction in predictions]
confusion_matrix = metrics.confusion_matrix(actual, predicted)
cm_display = metrics.ConfusionMatrixDisplay(confusion_matrix = confusion_matrix, display_labels = [1, -1])
cm_display.plot()
[14]:
<sklearn.metrics._plot.confusion_matrix.ConfusionMatrixDisplay at 0x7efd85b82200>
[15]:
test.Accuracy(test_labels,predictions)
[15]:
0.925